Statistics 2: Probability, Distributions, & Tests¶
Package/module refs:
- pandas for storing your data
- numpy also for storing data (as arrays), and other awesome things
- math.factorial for factorials
- scipy.stats for t-tests and distribution functions
- matplotlib.pyplot for visualizing your data
- my tutorial on plotting data. It’s focused on Astronomy but is universally (HA!) applicable
Handling Null Data¶
No dataset is perfect. Most data that you work with, especially
human-compiled data, will be somewhat incomplete. Missing values. When
incomplete data is loaded with
pandas.read_csv,
numpy.loadtxt,
or
numpy.genfromtxt,
those missing pieces will be filled by a default NaN (not a number)
value.
In [1]:
import pandas as pd
import numpy as np
incomplete_data = pd.read_csv("../downloads/nmhw_sample_data.csv")
incomplete_data
Out[1]:
| Name | Age | Hometown | Country | Desired Income | Favorite Food | Number of Cats | |
|---|---|---|---|---|---|---|---|
| 0 | James | 22 | New York | NaN | 75000.0 | NaN | 0.0 |
| 1 | Bob | 25 | Seattle | USA | 0.0 | pad thai | 2.0 |
| 2 | Annette | 28 | San Francisco | USA | NaN | spaghetti | 1.0 |
| 3 | Florence | 23 | Beijing | China | 60000.0 | xiaolong bao | 4.0 |
| 4 | Martha | 30 | Kansas City | NaN | NaN | corn | 3.0 |
| 5 | Desean | 27 | Newark | USA | 85000.0 | NaN | NaN |
| 6 | Jamal | 28 | New York | NaN | 65000.0 | pizza | 3.0 |
| 7 | Kaede | 31 | Kyoto | Japan | 65000.0 | sushi | 4.0 |
| 8 | Milton | 23 | Austin | NaN | 78000.0 | bbq | NaN |
| 9 | Ivana | 29 | Moscow | Russia | NaN | borscht | 1.0 |
| 10 | Jorge | 35 | Rio de Janeiro | Brasil | 90000.0 | farofa | 2.0 |
Always keep in mind that a “NaN” value *only* signifies a non-entry.
Often, it’s tempting to just replace NaN values with zeros, or
False, but to universally assume that you can or even should do that
doesn’t do justice to what a NaN is. To be sure, NaN values *are
numerical* but neither represent any actual number nor evaluate as
False, None, or infinity.
In [2]:
this_val = incomplete_data.Country.iloc[0]
print(this_val)
print(type(this_val))
print((this_val == 0) | (this_val < 0) | (this_val > 0) | (this_val == np.inf))
print((this_val == False) | (this_val == None))
if this_val:
print("This still prints something because it still represents a thing.")
nan
<class 'float'>
False
False
This still prints something because it still represents a thing.
There are many ways to handle NaN values and many entries (or
lackthereof) that should be recognized as such. Pandas allows you to
recognize a variety of values as null upon entry with the na_values
parameter of pandas.read_csv
In [3]:
incomplete_data = pd.read_csv("../downloads/nmhw_sample_data.csv", na_values=["null", "NULL", -9999, np.inf])
Once assigned to a variable, NaN values can be dealt with on a
column-by-column basis or throughout the entire DataFrame with the
fillna method. A NaN might be set to some default value, as you may
be able to assume a meaningful value for a non-entry. You may also
decide that a different column’s NaN may need to be evaluated in order
to get it out of the way in filtering or some other data process later
on.
In [4]:
incomplete_data.Country.fillna("USA")
Out[4]:
0 USA
1 USA
2 USA
3 China
4 USA
5 USA
6 USA
7 Japan
8 USA
9 Russia
10 Brasil
Name: Country, dtype: object
In [5]:
print(incomplete_data["Number of Cats"].fillna(-99))
0 0.0
1 2.0
2 1.0
3 4.0
4 3.0
5 -99.0
6 3.0
7 4.0
8 -99.0
9 1.0
10 2.0
Name: Number of Cats, dtype: float64
If your data is ordered, you can use the “method” parameter to fill NaNs
with the last seen value with method=ffill or with the next valid
data point method=bfill
In [6]:
incomplete_data.fillna(method="ffill")
Out[6]:
| Name | Age | Hometown | Country | Desired Income | Favorite Food | Number of Cats | |
|---|---|---|---|---|---|---|---|
| 0 | James | 22 | New York | NaN | 75000.0 | NaN | 0.0 |
| 1 | Bob | 25 | Seattle | USA | 0.0 | pad thai | 2.0 |
| 2 | Annette | 28 | San Francisco | USA | 0.0 | spaghetti | 1.0 |
| 3 | Florence | 23 | Beijing | China | 60000.0 | xiaolong bao | 4.0 |
| 4 | Martha | 30 | Kansas City | China | 60000.0 | corn | 3.0 |
| 5 | Desean | 27 | Newark | USA | 85000.0 | corn | 3.0 |
| 6 | Jamal | 28 | New York | USA | 65000.0 | pizza | 3.0 |
| 7 | Kaede | 31 | Kyoto | Japan | 65000.0 | sushi | 4.0 |
| 8 | Milton | 23 | Austin | Japan | 78000.0 | bbq | 4.0 |
| 9 | Ivana | 29 | Moscow | Russia | 78000.0 | borscht | 1.0 |
| 10 | Jorge | 35 | Rio de Janeiro | Brasil | 90000.0 | farofa | 2.0 |
In [7]:
incomplete_data.fillna(method="bfill")
Out[7]:
| Name | Age | Hometown | Country | Desired Income | Favorite Food | Number of Cats | |
|---|---|---|---|---|---|---|---|
| 0 | James | 22 | New York | USA | 75000.0 | pad thai | 0.0 |
| 1 | Bob | 25 | Seattle | USA | 0.0 | pad thai | 2.0 |
| 2 | Annette | 28 | San Francisco | USA | 60000.0 | spaghetti | 1.0 |
| 3 | Florence | 23 | Beijing | China | 60000.0 | xiaolong bao | 4.0 |
| 4 | Martha | 30 | Kansas City | USA | 85000.0 | corn | 3.0 |
| 5 | Desean | 27 | Newark | USA | 85000.0 | pizza | 3.0 |
| 6 | Jamal | 28 | New York | Japan | 65000.0 | pizza | 3.0 |
| 7 | Kaede | 31 | Kyoto | Japan | 65000.0 | sushi | 4.0 |
| 8 | Milton | 23 | Austin | Russia | 78000.0 | bbq | 1.0 |
| 9 | Ivana | 29 | Moscow | Russia | 90000.0 | borscht | 1.0 |
| 10 | Jorge | 35 | Rio de Janeiro | Brasil | 90000.0 | farofa | 2.0 |
You can choose to fill with the mean or median, however take note that strings can’t be averaged or median-ed. So be aware of what columns you’re fiddling with.
In [8]:
incomplete_data["Age"].fillna(incomplete_data.mean()["Age"])
Out[8]:
0 22
1 25
2 28
3 23
4 30
5 27
6 28
7 31
8 23
9 29
10 35
Name: Age, dtype: int64
In [9]:
incomplete_data["Desired Income"].fillna(incomplete_data.median()["Desired Income"])
Out[9]:
0 75000.0
1 0.0
2 70000.0
3 60000.0
4 70000.0
5 85000.0
6 65000.0
7 65000.0
8 78000.0
9 70000.0
10 90000.0
Name: Desired Income, dtype: float64
Other times it would make more sense to just leave NaNs as they are. Aggregate operations like means and sums will just ignore them. Though be aware that they will screw up iterative processes with strings.
In [10]:
print(incomplete_data["Desired Income"].mean())
incomplete_data["Favorite Food"].map(lambda x: x.split(" "))
64750.0
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-10-89f3d5c32e54> in <module>()
1 print(incomplete_data["Desired Income"].mean())
----> 2 incomplete_data["Favorite Food"].map(lambda x: x.split(" "))
/Users/Nick/anaconda/envs/python-dev-accelerator/lib/python3.5/site-packages/pandas/core/series.py in map(self, arg, na_action)
2119 index=self.index).__finalize__(self)
2120 else:
-> 2121 mapped = map_f(values, arg)
2122 return self._constructor(mapped,
2123 index=self.index).__finalize__(self)
pandas/src/inference.pyx in pandas.lib.map_infer (pandas/lib.c:63043)()
<ipython-input-10-89f3d5c32e54> in <lambda>(x)
1 print(incomplete_data["Desired Income"].mean())
----> 2 incomplete_data["Favorite Food"].map(lambda x: x.split(" "))
AttributeError: 'float' object has no attribute 'split'
It’s worth noting that while NaN values essentially mean the same no matter what, not all NaNs are created equal. A pandas nan is not the same as a numpy nan (although oddly-enough will be recognized as a nan-like value), and vice versa.
In [11]:
print(this_val == np.nan)
print(np.isnan(this_val))
print(pd.isnull(np.nan))
print(type(np.nan))
False
True
True
<class 'float'>
At the end of the day, no matter what you decide to do with NaNs make sure that you know your data and have an idea of what you want to do with it. All choices will follow from the nature of your data set and what your analysis entails.
Basic Rules of Probability¶
- The chance that some event happens given all events that could happen in that category.
- Probabilities denote the chance that something happens, not the guarantee
- \(0 \le p(x) \le 1\) or \(0\% \le p(x) \le 100\%\)
- For any set of associated but separate events, \(\displaystyle\sum_i p(x_i) = 1\)
- For related events A and B, the probablity that they happen together is
\(p(A \cap B) = p(A) \cdot p(B)\)
- The probability that either one of them happens is
\(p(A \cup B) = p(A) + p(B) - p(A \cap B)\)
Examples:¶
- p(coin toss = Head)
- p(rolling D20 and getting ≤ 11)
- p(5 coin tosses = Head)
- For bag of 4 white balls and 6 black balls, p(drawing 2 white then 2 black)
Probability Distributions¶
In [12]:
import matplotlib.pyplot as plt
%matplotlib inline
data = pd.read_csv("../downloads/prosperLoanData_truncated.csv", sep=",")
data = data[data.StatedMonthlyIncome < 2E4]
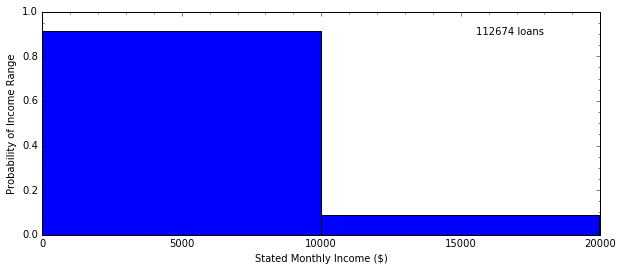
H, edges = np.histogram(data.StatedMonthlyIncome, bins=2)
plt.figure(figsize=(10, 4))
ax = plt.subplot(111)
ax.bar(edges[:-1], H / float(sum(H)), width=edges[1] - edges[0])
ax.text(0.9, 0.9, "%g loans" % len(data),
horizontalalignment="right", transform=ax.transAxes)
ax.set_xlabel("Stated Monthly Income ($)")
ax.set_ylabel("Probability of Income Range")
ax.minorticks_on()
plt.show()
In [13]:
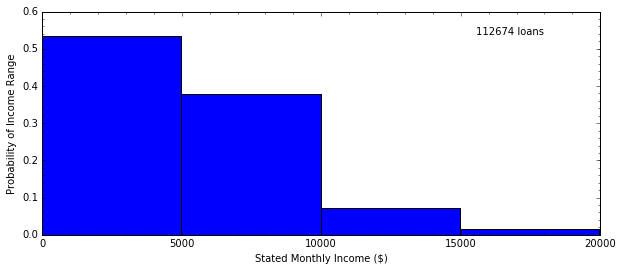
H, edges = np.histogram(data.StatedMonthlyIncome, bins=4)
plt.figure(figsize=(10, 4))
ax = plt.subplot(111)
ax.bar(edges[:-1], H / float(sum(H)), width=edges[1] - edges[0])
ax.text(0.9, 0.9, "%g loans" % len(data),
horizontalalignment="right", transform=ax.transAxes)
ax.set_xlabel("Stated Monthly Income ($)")
ax.set_ylabel("Probability of Income Range")
ax.minorticks_on()
plt.show()
In [14]:
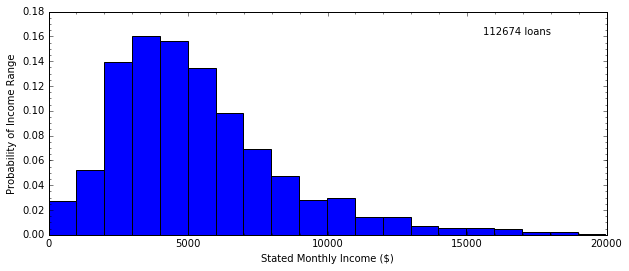
H, edges = np.histogram(data.StatedMonthlyIncome, bins=20)
plt.figure(figsize=(10, 4))
ax = plt.subplot(111)
ax.bar(edges[:-1], H / float(sum(H)), width=edges[1] - edges[0])
ax.text(0.9, 0.9, "%g loans" % len(data),
horizontalalignment="right", transform=ax.transAxes)
ax.set_xlabel("Stated Monthly Income ($)")
ax.set_ylabel("Probability of Income Range")
ax.minorticks_on()
plt.show()
In [15]:
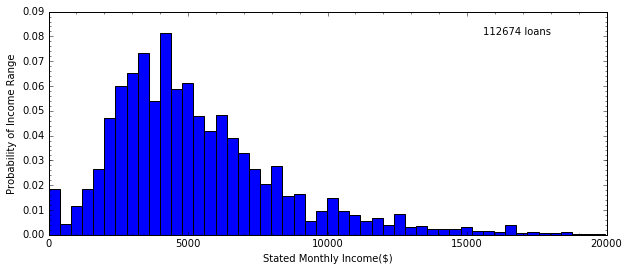
H, edges = np.histogram(data.StatedMonthlyIncome, bins=50)
plt.figure(figsize=(10, 4))
ax = plt.subplot(111)
ax.bar(edges[:-1], H / float(sum(H)), width=edges[1] - edges[0])
ax.text(0.9, 0.9, "%g loans" % len(data),
horizontalalignment="right", transform=ax.transAxes)
ax.set_xlabel("Stated Monthly Income($)")
ax.set_ylabel("Probability of Income Range")
ax.minorticks_on()
plt.show()
Note that here I use ax to access the plot space of the figure, so
that I can place text wherever I want it. If I don’t use the
transform parameter, then if I want to place text on a figure I have
to specify the exact coordinates in the data space for where I want it
to be. With transforming to the coordinate space of the figure, I can
use the fact that coordinate space goes from 0 to 1 horizontally and
vertically to place text easily.
One issue is that if you use ax, you set axis labels using
ax.set_xlabel and ax.set_ylabel instead of the normal methods
with plt.xlabel and plt.ylabel. Similar for setting axis limits.
Also note that with numpy arrays (and pandas DataFrames), you can perform arithmetic on the entire object at once. Also note that with numpy arrays specifically, you cannot alter the size of the array once it’s been created. Fixed-size arrays make it faster, but the immutability of size can be a little difficult. At least you can alter the values inside the array easily through indexing.
Cumulative Probability Distribution¶
- The probability of getting a value less than x
In [16]:
H, edges = np.histogram(data.StatedMonthlyIncome, bins=np.linspace(0, 2E4, 81))
incomes = [5000, 10000, 15000]
fig = plt.figure(figsize=(10, 6))
fig.subplots_adjust(hspace=0) # smooshes vertical subplots together
ax = plt.subplot(211) # 2 rows, 1 column, put this figure in the first cell
ax.bar(edges[:-1], H / float(sum(H)), width=edges[1] - edges[0])
for ii in incomes:
ax.plot([ii, ii], [0, 0.06], linestyle="--", color='k')
ax.set_ylabel("Probability of Income Range")
ax.set_xticklabels([]) # kills the numbers on the x-axis for this plot
ax.minorticks_on()
ax = plt.subplot(212) # 2 rows, 1 column, put this figure in the first cell
probabilities = H.cumsum() / float(sum(H))
ax.plot(edges[:-1], probabilities, linewidth=5, color="r")
for ii in incomes:
prob = probabilities[edges[:-1] == ii]
ax.plot([ii, ii], [0, prob], linestyle="--", color='k')
ax.plot([0, ii], [prob, prob], linestyle="--", color='k')
ax.text(ii + 2.5E2, prob * 0.5, "p(x): %.2f" % prob)
ax.set_xlabel("Stated Monthly Income ($)")
ax.set_ylabel("Cumulative Probability")
ax.set_ylim(0, 0.99)
ax.minorticks_on()
plt.savefig("../downloads/cumulative_distribution.png")
plt.show()
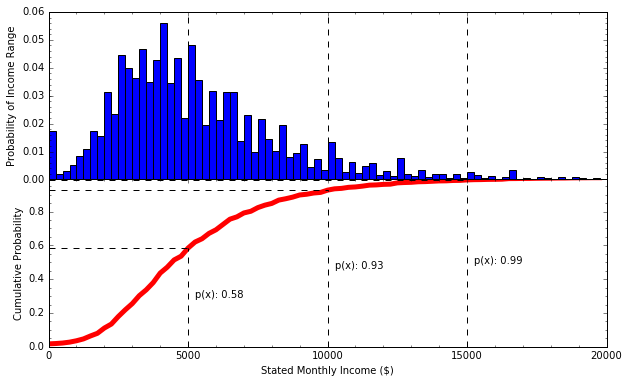
Numpy arrays have several methods, one of which is cumsum(). This
method performs a cumulative sum on your array from left to right.
arr = np.array([1, 1, 1, 1, 1, 1, 1, 1])
arr.cumsum()
>>> array([1, 2, 3, 4, 5, 6, 7, 8])
- “If I choose a person from this data set at random, there’s a 58% chance that their monthly salary is less than $5,000”
- ”..., there’s a 35% chance their monthly salary is between $5,000 and $10,000”
- ”..., there’s a 1% chance their monthly salary is greater than $15,000”
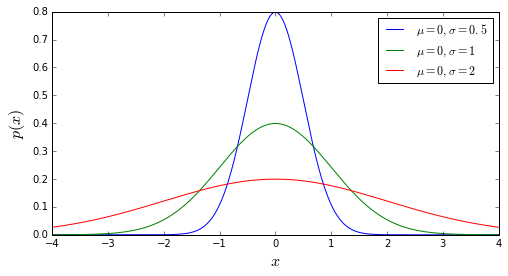
The Gaussian/Normal Distribution¶
- Theoretical curve describing the probability of getting some value x, given the mean and standard deviation of the data
- \(p(x|\sigma, \mu) = \displaystyle\frac{1}{\sqrt{2\pi\sigma^2}}\exp{\left[\frac{-(x - \mu)^2}{2\sigma^2}\right]}\)
- The most commonly-used distribution
- Ref: Central Limit Theorem
- The Gist: The mean of samples drawn from an almost arbitrary distribution will approx. follow a Gaussian
- Applied to everything, even when not appropriate
In [17]:
del data
def gaussian(x, mu, std):
return (1. / np.sqrt(2 * np.pi * std**2) * np.exp(-0.5 * ((x - mu) / std)**2))
x = np.linspace(-4, 4, 1000)
fig = plt.figure(figsize=(8, 4))
plt.plot(x, gaussian(x, 0, 0.5), label="$\mu = 0, \sigma = 0.5$")
plt.plot(x, gaussian(x, 0, 1), label="$\mu = 0, \sigma = 1$")
plt.plot(x, gaussian(x, 0, 2), label="$\mu = 0, \sigma = 2$")
plt.xlabel("$x$", fontsize=16)
plt.ylabel("$p(x)$", fontsize=16)
plt.legend()
plt.savefig("../downloads/gaussian_distribution.png")
plt.show()
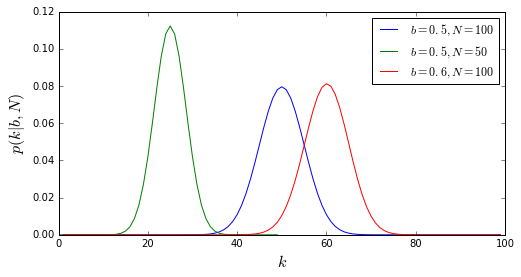
The Binomial Distribution¶
- Describes the distribution of a variable that can only take two values (0/1, False/True, Heads/Tails)
- For N trials...
- I know the probability b of getting one outcome per trial...
- I can describe the probability of getting k successes of that outcome:
- The mean \(\bar{k} = b\,N\)
- The standard deviation \(\sigma_k = \sqrt{N \,b \,(1 - b)}\)
- NOTE: does not describe probability of getting consecutive successes or any particular combination of successes & failures
In [18]:
from math import factorial
def binomial(k, N, b):
return float(factorial(N)) / (factorial(k) * factorial(N - k)) * b ** k * (1 - b)**(N - k)
x = range(1, 100)
y0 = [binomial(val, 100, 0.5) for val in x]
y1 = [binomial(val, 50, 0.5) for val in range(0, 50)]
y2 = [binomial(val, 100, 0.6) for val in x]
fig = plt.figure(figsize=(8, 4))
plt.plot(x, y0, label="$b = 0.5, N = 100$")
plt.plot(range(0, 50), y1, label="$b = 0.5, N = 50$")
plt.plot(x, y2, label="$b = 0.6, N = 100$")
plt.xlabel("$k$", fontsize=16)
plt.ylabel("$p(k | b, N)$", fontsize=16)
plt.legend()
plt.savefig("../downloads/binomial_distribution.png")
plt.show()
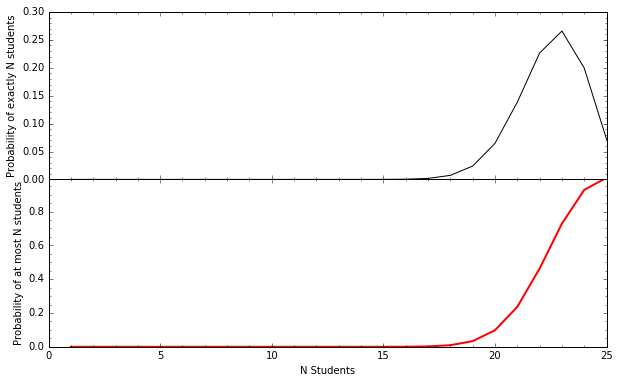
Example: the probability of \(\ge k\) Code Fellows students getting a job after 6 months¶
In [19]:
from scipy.stats import binom
n_students = np.arange(1, 26)
total_students = 25
prob_of_success = 0.9
individual_probability = [binom.pmf(x, total_students, prob_of_success) for x in n_students]
cumulative_probability = [binom.cdf(x, total_students, prob_of_success) for x in n_students]
fig = plt.figure(figsize=(10, 6))
fig.subplots_adjust(hspace=0)
ax = plt.subplot(211)
ax.plot(n_students, individual_probability, color="k")
ax.set_xticklabels([])
ax.set_ylabel("Probability of exactly N students")
ax.minorticks_on()
ax = plt.subplot(212)
ax.plot(n_students, cumulative_probability, color="r", linewidth=2)
ax.set_xlabel("N Students")
ax.set_ylabel("Probability of at most N students")
ax.set_ylim(0, 0.99)
ax.minorticks_on()
plt.show()
Hypothesis Testing and Measurement Significance¶
- Chance occurrence vs meaningful occurrence
- Sticking with the Gaussian distribution of random samples
- Either reject the Null Hypothesis \(H_0\) or fail to reject it.
- Can never PROVE absolutely True or absolutely False
- Hypothesis conclusions entirely dependent on the sample. Cannot be assumed to be universal.
- Significance levels (\(\alpha\)) help decide whether to reject
\(H_0\)
- Typical \(\alpha\)-levels: 0.05, 0.025, 0.01
- Statistical tests produce two things:
- a numerical statistic showing the distance between the means of two distributions (more later)
- a “p-value” showing the probability of getting this result given the null hypothesis
- If your p-value is less than some alpha-level, then it’s
statistically significant to that level
- ex: p = 0.035; my test rejects the null hypothesis at \(\alpha = 0.05\) significance level, but not at the \(\alpha = 0.025\) level.
- ex: p = 0.13; my tests fails to reject the null hypothesis
Students’ t-Test¶
“Students” was the pseudonym of the originator
Computes a statistic (the “t” statistic) based on the means and variances of samples
\[t = \displaystyle\frac{\bar{x_1} - \bar{x_2}}{\sqrt{\frac{s_1^2}{n_1} + \frac{s_2^2}{n_2}}}\]Defs:
- \(s_i\): standard deviation of sample \(i\)
- \(n_i\): number of items in sample \(i\)
Use Cases
- test whether a subsample is significantly different from the population
- test if two subsamples from the same population are significantly different
- test if two independent samples are significantly different
Note: Data should be somewhat “normally” distributed (so visualize). Non-normal data needs a different test (next time
t-Test example: Seattle Weather in 2010¶
- Hypothesis: The temperature during the Spring was significantly different than the temperature during the Fall
- Null hypothesis: The temperature during the Spring wasn’t significantly different
- Define terms:
- Spring: March through May
- Fall: September through November
In [20]:
import os
data_dir = "../downloads/seattle_weather/"
weather_files = [
data_dir + f for f in os.listdir(data_dir) if f.startswith("MonthlyHistory")]
seattle_weather = pd.read_csv(weather_files[0])
for f in weather_files[1:]:
indata = pd.read_csv(f)
if "PDT" in indata.columns:
indata.rename(columns={'PDT': 'PST'}, inplace=True)
seattle_weather = pd.concat((seattle_weather, indata), ignore_index=True)
pandas.concat takes two DataFrames and merges them into one
DataFrame. If you don’t set ignore_index=True then it will preserve
the index values that each individual DataFrame had before merging,
which can become problematic when slicing the new merged frame
Note that when concatenating DataFrames, any columns that exist in one and not in the other will get filled with null values. Above, I just change the name of the “PDT” column because not every data file has that column despite them all showing roughly the same data.
In [21]:
seattle_weather["year"] = seattle_weather.PST.map(lambda x: float(x.split("-")[0]))
seattle_weather["month"] = seattle_weather.PST.map(lambda x: float(x.split("-")[1]))
seattle_weather["day"] = seattle_weather.PST.map(lambda x: float(x.split("-")[2]))
You can create new columns in a DataFrame much like how you can create
new keys in a dictionary, by specifying the column name that you want.
You can also get data from columns in two ways: calling that column up
with the square bracket notation DataFrame["column_name"] or with
dot notation DataFrame.column_name. The main difference is that dot
notation cannot handle column names with spaces.
In [22]:
spring_2010 = (seattle_weather.year == 2010) & (seattle_weather.month > 2) & (seattle_weather.month < 6)
fall_2010 = (seattle_weather.year == 2010) & (seattle_weather.month > 8) & (seattle_weather.month < 12)
You can select indices of data matching specific criteria. The above
lines produce two pandas Series objects (effectively a one-column
DataFrame) holding boolean values of True when a row matches your
criteria, and False when it doesn’t. You can do the same with numpy
arrays (where the same selection will produce numpy arrays filled with
booleans)
In [23]:
plt.figure(figsize=(10, 4))
opacity = 0.5
mean_temp = "Mean TemperatureF"
plt.hist(seattle_weather[spring_2010][mean_temp], bins=np.arange(20, 76, 4), alpha=opacity, label="Spring 2010")
plt.hist(seattle_weather[fall_2010][mean_temp], bins=np.arange(20, 76, 4), alpha=opacity, label="Fall 2010")
plt.legend()
plt.xlabel("Mean Temperature (F)")
plt.ylabel("N days")
plt.show()
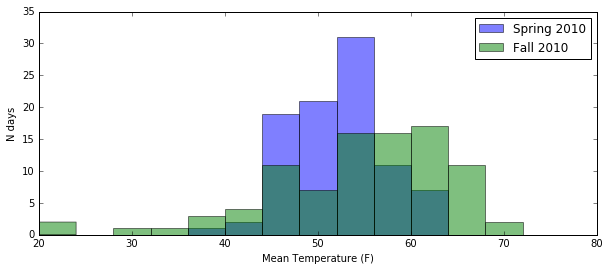
The alpha parameter controls the opacity of whatever you’re
plotting, whether they are bars, points, lines, circles, whatever.
Opacity goes from 0 to 1. Works best when you you have heavily
overlapping data. In the event of overlapping data, plot the larger data
set first (in the background) so that it doesn’t drown out the smaller
set.
In [24]:
from scipy.stats import ttest_ind
t_stat, p_value = ttest_ind(seattle_weather[spring_2010][mean_temp],
seattle_weather[fall_2010][mean_temp])
print("Results:\n\tt-statistic: %.5f\n\tp-value: %.5f" % (t_stat, p_value))
Results:
t-statistic: -2.09063
p-value: 0.03796
I can reject the null hypothesis at the p < 0.05 significance level. At that level, the mean temperature in Spring and Fall 2010 was significantly different.
t-tests in SciPy.stats¶
- `ttest_ind <http://docs.scipy.org/doc/scipy-0.16.0/reference/generated/scipy.stats.ttest_ind.html#scipy.stats.ttest_ind>`__: performs a t-test on two sample distributions and returns the t-statistic (useful later) and the p-value; useful for two independent samples from the same population (or very similar but different populations)
- `ttest_1samp <http://docs.scipy.org/doc/scipy-0.16.0/reference/generated/scipy.stats.ttest_1samp.html#scipy.stats.ttest_1samp>`__: useful for one sample distribution in comparison to the mean of a population
- `ttest_rel <http://docs.scipy.org/doc/scipy-0.16.0/reference/generated/scipy.stats.ttest_rel.html#scipy.stats.ttest_rel>`__: useful for two related samples
